Feature Description
High/Low Clustering measures the degree of clustering for either high values or low values using the Getis-Ord General G statistic. The General G Index is an inferential statistic that uses limited data to estimate characteristics of the entire population. When the returned P-value is small and statistically significant, we can reject the null hypothesis. In such cases, a positive Z-score indicates the observed General G Index is larger than expected, suggesting clustering of high attribute values in the study area; a negative Z-score indicates the observed General G Index is smaller than expected, suggesting clustering of low attribute values.
Application Cases
- Detecting abnormal peaks in emergency room visits may indicate outbreaks of local or regional health issues.
- Comparing spatial patterns of different retail industries in urban areas to understand competitive sectors (e.g., car dealerships) and non-competitive sectors (e.g., health centers/gyms) through comparative shopping analysis.
- Assessing the degree of spatial clustering over time to examine changes, such as analyzing urban population clustering patterns during urban development and densification processes.
Feature Entry
- Spatial Statistics Tab -> Analyzing Patterns -> High/Low Clustering.
- Toolbox -> Spatial Statistics -> Analyzing Patterns -> High/Low Clustering.
Parameter Description
- Source Dataset: The vector dataset to analyze, supporting point, line, and polygon datasets.
- Evaluation Field: The numeric field representing attribute values for analysis.
- Conceptualization Model: Defines spatial relationships between features. More realistic models yield more accurate results.
- Fixed Distance: Suitable for point data and polygon data with varying sizes.
- Polygon Adjacent (Common Edges/Intersect): For polygons sharing edges or intersections.
- Polygon Adjacent (Node/Common Edges/Intersect): For polygons sharing nodes, edges, or intersections.
- Inverse Distance: All features influence each other with weights inversely proportional to distance, suitable for continuous data.
- Inverse Distance Square: Similar to inverse distance but with faster distance decay (weights = 1/distance²).
- K Nearest Neighbors: Uses K nearest features with equal weights (1). Effective for ensuring minimum neighbors and handling uneven distributions.
- Spatial Weight Matrix: Uses a spatial weight matrix file (.swmb) to model spatial relationships, ideal for network-based analyses like crime cluster detection.
- Undifferentiated Region: Combines fixed distance and inverse distance models. Features within a threshold have equal weights; others follow inverse distance rules.
- Distance Threshold: "-1" calculates default distance; "0" treats all as neighbors; positive values define adjacency range.
- Inverse Distance Power: Controls distance decay rate. Higher values reduce distant influences faster.
- Number of Neighbors: Specifies K for K Nearest Neighbors model.
- Distance Measurement: Euclidean Distance or Manhattan Distance. Details in Spatial Statistics Glossary.
- Spatial Weights Matrix Standardization: Normalizes weights by row sums (0-1 range), recommended for administrative boundaries to reduce bias from varying neighbor counts.
Result Interpretation
Analysis results are stored in a CAD dataset and displayed on the map.
The High/Low Clustering analysis returns five parameters: General G Index, Expected Value, Variance, Z-score, and P-value. As an inferential statistic, a small P-value (<0.05) allows rejecting the null hypothesis. A positive Z-score indicates high-value clustering; a negative Z-score indicates low-value clustering:
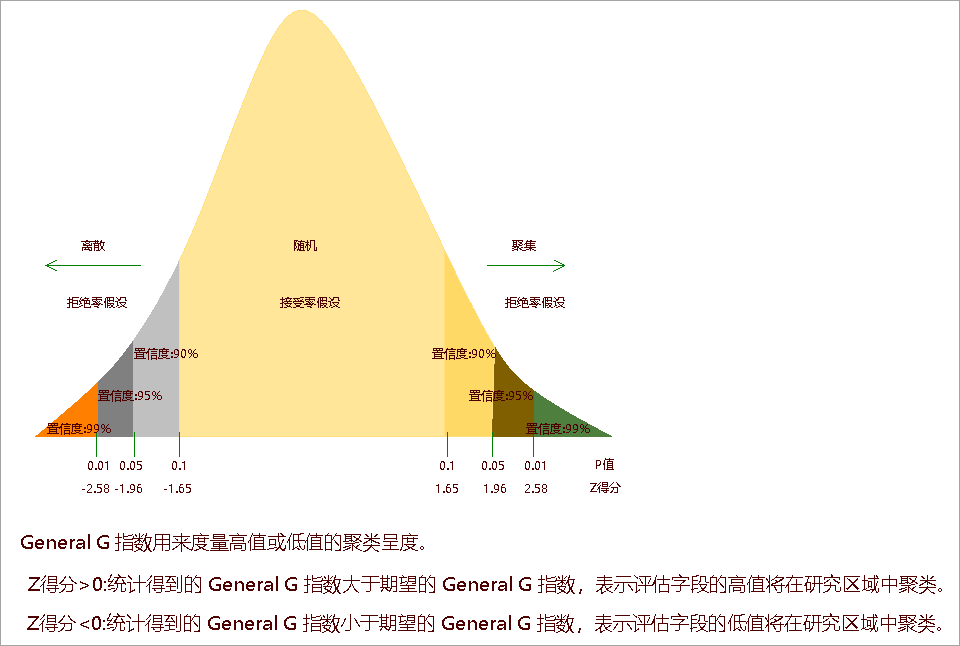
Use High/Low Clustering to detect spatial peaks of high values in uniformly distributed data. When high and low clusters cancel out (observed G ≈ expected G), consider Spatial Autocorrelation analysis instead.
Example
Sample Data: Download case data (requires extraction after download).
Analyzing viral hepatitis incidence using: Evaluation Field = case count, Conceptualization Model = inverse distance, Distance Measurement = Euclidean, Standardized spatial weights. Results show:

Conclusions: P-value <0.01 and Z-score >2.58 indicate 99% confidence in significant high-value clustering of hepatitis cases.
Related Topics
Incremental Spatial Autocorrelation



